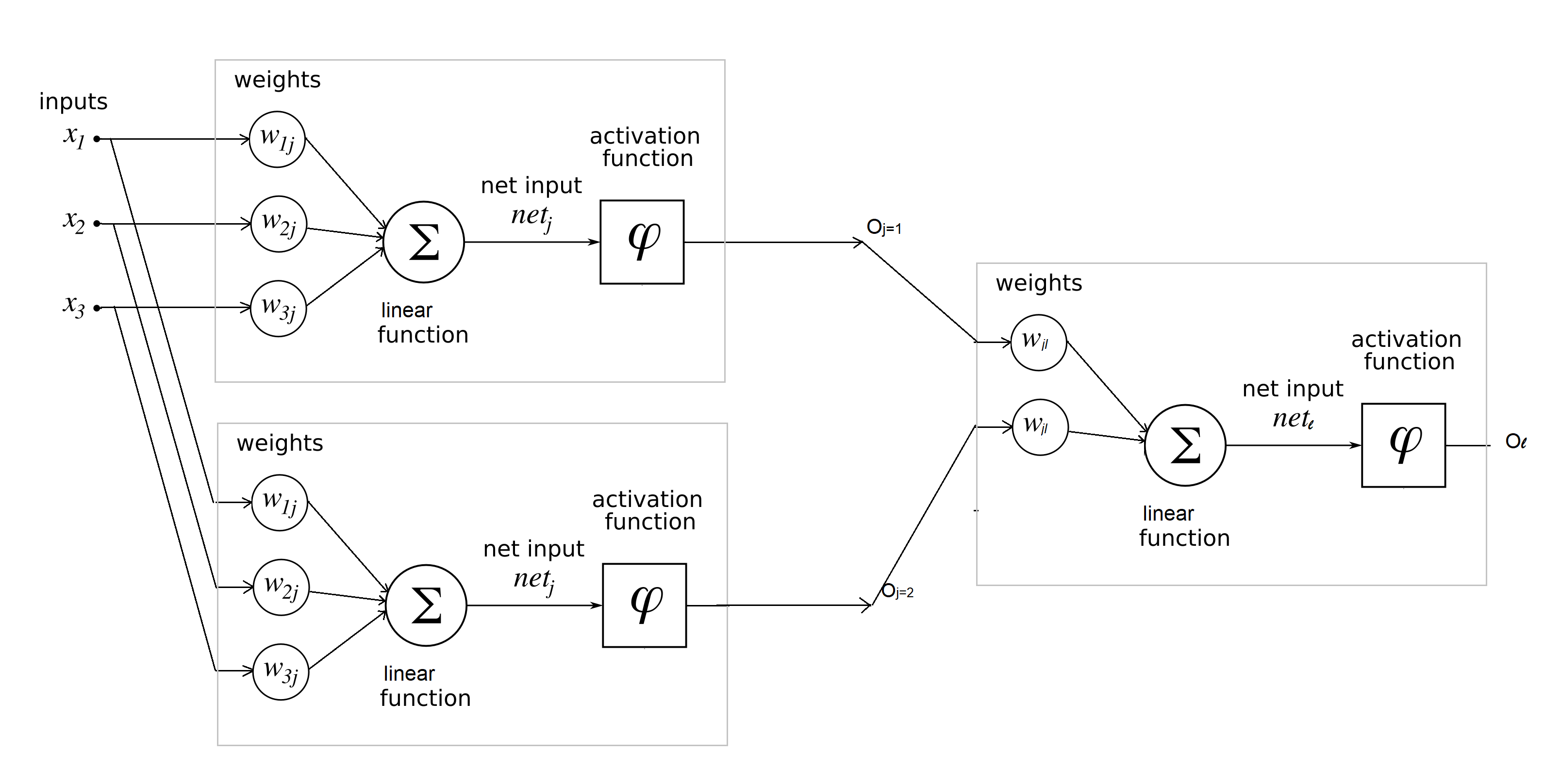
Building neural network step-by-step in raw python¶
1. one single neuron for linear regression:¶
The neuron is just two matrices, one of intercepts, the other one of bias. if we have m inputs / features / independent variables $\mathbf{x}=[x_1,\cdots,x_m]^T$, we can write the neuron's function as a linear regression: $y=\mathbf{w}^T \mathbf{x}+b$, where intercepts are $\mathbf{w}=[w_1,\cdots,w_m]^T$.
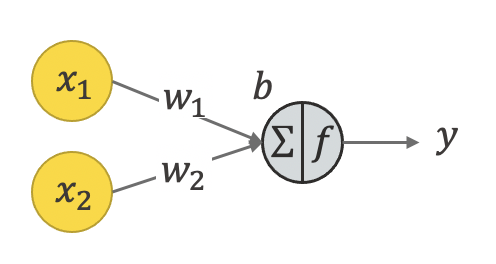
2. one single neuron for non-linear regression:¶
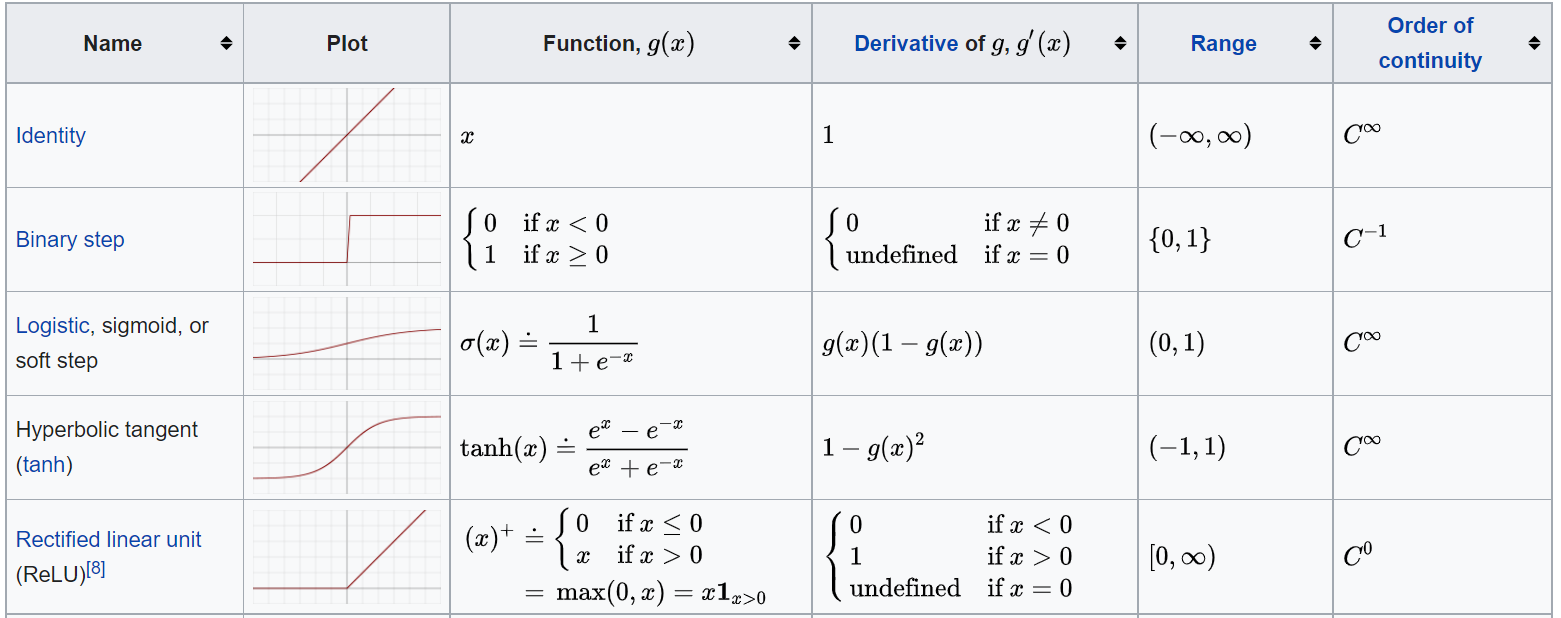
The neuron can also be for non-linear regression, through a non-linear function called the activation function. This function maps the $y$ into $y_{\text{hat}}$ within a certain range. Choice of the function is such that 1) the neuron can act like a switch: when y is below the threshold value, the neuron is turned off; otherwise it's turned on; 2) it is better to use a smooth function so that we can use its derivative for learning. The idea of gradient descent for learning is the same as what we discussed in the previous courses.
A typical activation function can be for example sigmoid function: $y =\cfrac{1}{1+exp(-x)}$. Some others are https://en.wikipedia.org/wiki/Activation_function: